library("SBC")
library("brms")
library("cmdstanr")
library("ggplot2")
library("colorspace")
library("cowplot")
library("future")Intro
Relevant parts of the code given here is inspired by the resources of the great SBC package (Modrák et al., 2023) published at https://hyunjimoon.github.io/SBC/index.html.
Also, coding of posterior p-values is inspired by the split predictive check (Li & Huggins, 2024) R code published at https://github.com/TARPS-group/split-predictive-checks.
Software
We use the statistical software environment R (R Core Team, 2024), and R add-on packages colorspace (Stauffer et al., 2009), SBC (Modrák et al., 2023), brms (Bürkner, 2017, 2018, 2021), cmdstanr (Gabry et al., 2024), ggplot2 (Wickham, 2016), and future (Bengtsson, 2021).
This document is produced using Quarto (Allaire et al., 2024).
Organize R Session
Load packages:
Settings:
rm(list = ls())
options(brms.backend = "cmdstanr")
# Using parallel processing
plan(multisession)
options(SBC.min_chunk_size = 5)
# Setup caching of results
cache_dir <- "./_SBC_cache"
if(!dir.exists(cache_dir)) {
dir.create(cache_dir)
}Data preparation function for graphical uniformity checks:
## from https://github.com/TeemuSailynoja/simultaneous-confidence-bands/tree/main
dat_ecdf_diff <- function (x, variables = NULL, K = NULL,
gamma = NULL, prob = 0.95,
combine_variables = NULL,
ecdf_alpha = NULL, ...,
parameters = NULL) {
if (!is.null(parameters)) {
warning("The `parameters` argument is deprecated use `variables` instead.")
if (is.null(variables)) {
variables <- parameters
}
}
ecdf_data <- data_for_ecdf_plots(x, variables = variables,
prob = prob, K = K, gamma = gamma, combine_variables = combine_variables,
ecdf_alpha = ecdf_alpha, ...)
if (ecdf_data$N < 50 && is.null(K)) {
message("With less than 50 simulations, we recommend using plot_ecdf as it has better fidelity.\n",
"Disable this message by explicitly setting the K parameter. ",
"You can use the strings \"max\" (high fidelity) or \"min\" (nicer plot) or choose a specific integer.")
}
N <- ecdf_data$N
K <- ecdf_data$K
z <- ecdf_data$z
ecdf_df <- dplyr::mutate(ecdf_data$ecdf_df, z_diff = ecdf -
z, type = "sample ECDF")
limits_df_trans <- dplyr::mutate(ecdf_data$limits_df, ymax = upper -
uniform_val, ymin = lower - uniform_val, type = "theoretical CDF")
return(list('ecdf_df' = ecdf_df,
'limits_df_trans' = limits_df_trans))
}Data Generator
The data can be generated using the following code – note that we need to be careful to match the parameter names as brms uses them.
f_generate_y <- function(sim_df,
b_Intercept,
b_x,
grouping_term, ## sd_g__Intercept or r_g
sigma) {
if (is.matrix(grouping_term)) {
r_g <- grouping_term
} else {
G <- length(levels(sim_df$g))
r_g <- matrix(rnorm(n = G, mean = 0, sd = grouping_term),
nrow = G, ncol = 1,
dimnames = list(levels(sim_df$g), "Intercept"))
}
sim_df$'mu' <- exp(b_Intercept +
sim_df$'x' * b_x +
r_g[sim_df$'g'])
sim_df$'sigma' <- sigma
sim_df$'y' <- rnorm(n = nrow(sim_df),
mean = sim_df$'mu',
sd = sim_df$'sigma')
return(sim_df)
}
f_generate_one_sim <- function(N, G) {
prior_mu <- list('b_Intercept' = 0,
'b_x' = 1,
'sd_g__Intercept' = .5,
'sigma' = 1)
prior_sd <- list('b_Intercept' = .1,
'b_x' = .1,
'sd_g__Intercept' = .1,
'sigma' = .1)
b_Intercept <- rnorm(n = 1,
mean = prior_mu$b_Intercept,
sd = prior_sd$b_Intercept)
b_x <- rnorm(n = 1,
mean = prior_mu$b_x,
sd = prior_sd$b_x)
sd_g__Intercept <- rnorm(n = 1,
mean = prior_mu$sd_g__Intercept,
sd = prior_sd$sd_g__Intercept)
sd_g__Intercept <- abs(sd_g__Intercept)
sigma <- rnorm(n = 1,
mean = prior_mu$sigma,
sd = prior_sd$sigma)
sigma <- abs(sigma)
indx <- rep(c(5, 25, 45), c(6, 8, 6))
if (sum(indx) != 500) {
stop("'N' must be 500 currently!")
}
g <- paste0("Group ", formatC(1:G, width = 2, flag = "0"))
g <- sample(g)
g <- rep(g, indx)
g <- as.factor(g)
r_g <- matrix(rnorm(n = G, mean = 0, sd = sd_g__Intercept),
nrow = G, ncol = 1,
dimnames = list(levels(g), "Intercept"))
sim_df_skeleton <- data.frame('x' = 2 * runif(N),
'g' = g)
return(list('variables' = list('b_Intercept' = b_Intercept,
'b_x' = b_x,
'sd_g__Intercept' = sd_g__Intercept,
'r_g' = r_g,
'sigma' = sigma),
'generated' = f_generate_y(sim_df = sim_df_skeleton,
b_Intercept,
b_x,
grouping_term = r_g,
sigma)))
}
f_generate_n_sims <- SBC_generator_function(f_generate_one_sim,
N = 500,
G = 20)
set.seed(0)
datasets <- generate_datasets(f_generate_n_sims, 100)
head(datasets$generated[[1]]) x g mu sigma y
1 0.6933670 Group 11 2.010588 1.127243 1.257388
2 0.6675499 Group 11 1.960995 1.127243 3.026448
3 0.9527025 Group 11 2.583906 1.127243 3.072794
4 1.7843967 Group 11 5.776880 1.127243 6.909938
5 1.7286789 Group 11 5.473749 1.127243 5.033990
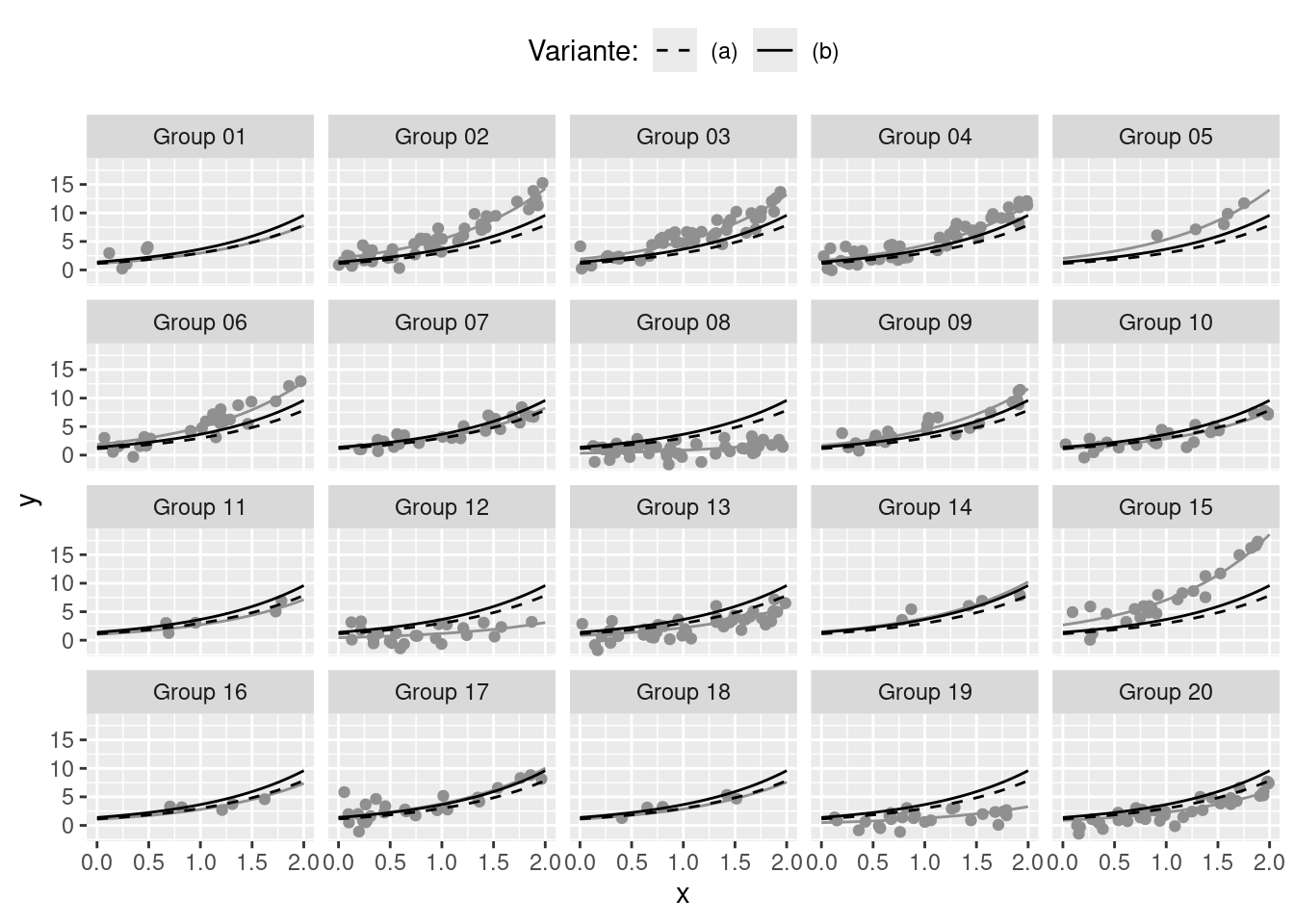
6 0.7799791 Group 14 3.143628 1.127243 3.567889Descriptive Plot
r <- 1
dat <- as.data.frame(expand.grid(x = seq(0, 2, by = .05), g = 1:20))
b <- as.numeric(datasets$variables[r, ])
dat$y <- exp(b[1] + dat$x * b[2] + b[dat$g + 3])
dat$g <- paste0("Group ", formatC(dat$g, width = 2, flag = "0"))
dat$k <- dat$l <- "Single group"
dat <- rbind(dat,
data.frame(x = seq(0, 2, by = .05),
y = exp(b[1] + dat$x * b[2]),
g = "Group 21",
k = "(a)",
l = "Average"),
data.frame(x = seq(0, 2, by = .05),
y = exp(b[1] + dat$x * b[2] + .5 * b[3]^2),
g = "Group 22",
k = "(b)",
l = "Average"))
sdat <- subset(dat, l == "Average")
sdat$g <- NULL
paint <- colorspace::sequential_hcl(n = 20, pal = "Grays")[10]
ggplot(data = datasets$generated[[r]], aes(x = x, y = y)) +
geom_line(data = subset(dat, l != "Average"), color = paint) +
geom_point(, color = paint) +
geom_line(data = sdat, aes(group = k, linetype = k), color = 1) +
scale_linetype_manual(values = c(2, 1)) +
facet_wrap(~ g) +
labs(linetype = "Variante:") +
theme(legend.position = "top")Backend
priors <-
prior(normal(0,.1), class = "b", coef = "Intercept") +
prior(normal(1,.1), class = "b", coef = "x") +
prior(normal(.5,.1), class = "sd") +
prior(normal(1,.1), class = "sigma")
stan_file_filename <- "brms_log_link_linear_mixed_model.stan"
f_backend <- SBC_backend_brms(bf(y ~ 0 + Intercept + x + (1 | g)),
family = gaussian(link = "log"),
prior = priors,
chains = 1,
template_data = datasets$generated[[1]],
out_stan_file = file.path(cache_dir,
stan_file_filename))
log_lik_log_link_lmm <- derived_quantities(
log_lik = sum(dnorm(y,
mean = exp(b_Intercept + x * b_x + r_g[g]),
sd = sigma,
log = TRUE)))
results <- compute_SBC(datasets,
f_backend,
dquants = log_lik_log_link_lmm,
cache_mode = "results",
cache_location = file.path(cache_dir,
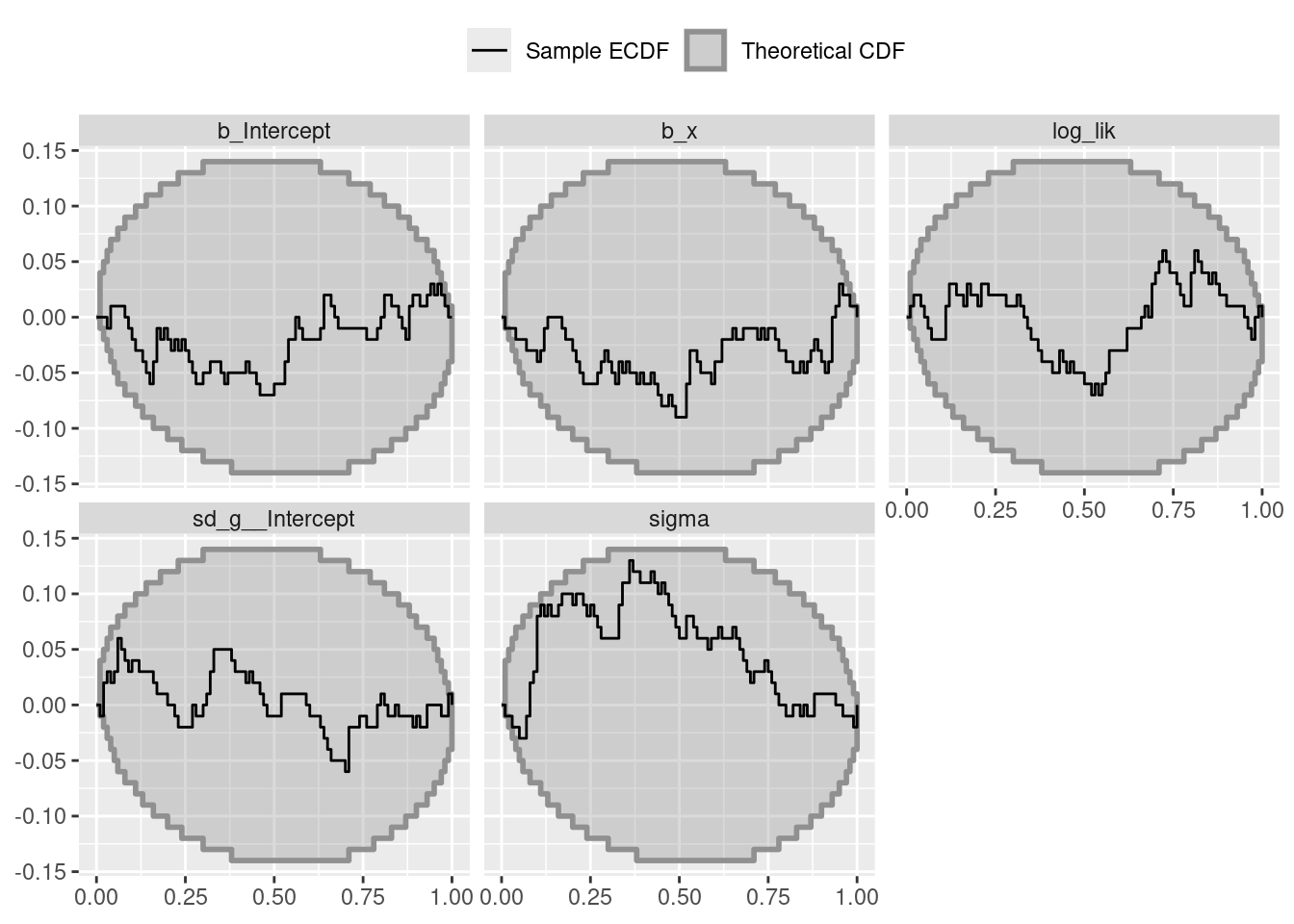
"results_log_link_linear_mixed_model"))tmp <- results$result$stats[, c("sim_id", "variable", "rank")]
tmp <- dat_ecdf_diff(tmp,
variables = c("b_Intercept", "b_x", "sd_g__Intercept", "sigma",
"log_lik"),
max_rank = 99)
tmp$variable <- factor(tmp$variable,
levels = c("b_Intercept", "b_x", "sd_g__Intercept", "sigma",
"log_lik"))
col_color <- c(colorspace::sequential_hcl(n = 20, pal = "Grays")[10], "black")
col_fill <- c(colorspace::sequential_hcl(n = 20, pal = "Grays")[10], "black")
names(col_color) <- names(col_fill) <-
c("theoretical CDF", "sample ECDF")
l <- c('theoretical CDF' = expression("Theoretical CDF"),
'sample ECDF' = expression("Sample ECDF"))ggplot(tmp$ecdf_df, aes(color = type, fill = type)) +
geom_ribbon(data = tmp$limits_df_trans,
aes(x = x, ymax = ymax, ymin = ymin), alpha = .33,
linewidth = 1) +
geom_step(aes(x = z, y = z_diff, group = variable)) +
scale_color_manual(name = "", values = col_color, labels = l) +
scale_fill_manual(name = "", values = col_fill, labels = l) +
scale_alpha_identity() +
xlab(NULL) +
ylab(NULL) +
facet_wrap(~ variable) +
theme(strip.text = element_text(margin = margin(b = 2, t = 2))) +
theme(legend.position = "top")Calculate QOI-Check
Preparations:
f_predict_brms_gaussian <- function(fit, nd) {
predict(fit, newdata = nd,
summary = F,
allow_new_levels = T,
sample_new_levels = "gaussian")
}
f_predict_brms_uncrtnty <- function(fit, nd) {
predict(fit, newdata = nd,
summary = F,
allow_new_levels = T,
sample_new_levels = "uncertainty")
}
set.seed(123456789)
p_stat_mean_ref_gr_gaussian_a <- p_stat_mean_ref_gr_uncrtnty_a <-
p_stat_mean_ref_gr_gaussian_b <- p_stat_mean_ref_gr_uncrtnty_b <-
p_stat_mean_sim_df_gaussian_a <- p_stat_mean_sim_df_uncrtnty_a <-
p_stat_mean_sim_df_gaussian_b <- p_stat_mean_sim_df_uncrtnty_b <-
## [October 22, 2024]
p_stat_mean_ref_gr_gaussian_c <- p_stat_mean_ref_gr_uncrtnty_c <-
p_stat_mean_ref_gr_gaussian_d <- p_stat_mean_ref_gr_uncrtnty_d <-
p_stat_mean_sim_df_gaussian_c <- p_stat_mean_sim_df_uncrtnty_c <-
p_stat_mean_sim_df_gaussian_d <- p_stat_mean_sim_df_uncrtnty_d <-
##
p_stat_mean_ref_gr_gaussian_e <- p_stat_mean_ref_gr_uncrtnty_e <-
p_stat_mean_ref_gr_gaussian_f <- p_stat_mean_ref_gr_uncrtnty_f <-
p_stat_mean_sim_df_gaussian_e <- p_stat_mean_sim_df_uncrtnty_e <-
p_stat_mean_sim_df_gaussian_f <- p_stat_mean_sim_df_uncrtnty_f <-
##
p_stat_mean_eq_a_a <- p_stat_mean_eq_a_b <- p_stat_mean_eq_a_c <-
p_stat_mean_eq_a_d <- p_stat_mean_eq_a_e <- p_stat_mean_eq_a_f <-
##
p_stat_mean_eq_b_a <- p_stat_mean_eq_b_b <- p_stat_mean_eq_b_c <-
p_stat_mean_eq_b_d <- p_stat_mean_eq_b_e <- p_stat_mean_eq_b_f <-
##
rep(NA, length(results$fits))Loop through the simulation runs and calculate all QOI variants for each run:
for (r in 1:length(results$fits)) {
## Prior in repetition r:
prior_r <- subset(results$stats, sim_id == r)
## Methods a and b:
equation_a <- exp(subset(prior_r, variable == "b_Intercept")$simulated_value +
1 * subset(prior_r, variable == "b_x")$simulated_value)
equation_b <- exp(subset(prior_r, variable == "b_Intercept")$simulated_value +
1 * subset(prior_r, variable == "b_x")$simulated_value +
.5 * (subset(prior_r,
variable == "sd_g__Intercept")$simulated_value^2))
## Reference grid data 'ref_gr':
g <- paste0("g", formatC(101:300, width = 3, flag = "0"))
ref_gr <- data.frame('x' = 1,
'g' = as.factor(g))
## Duplicate observation data 'sim_df':
sim_df <- results$fits[[r]]$data[, c('x', 'g')]
sim_df$x <- 1 ## November 15, 2024
g <- 100 + as.numeric(sim_df$g) ## 'break' original levels
g <- paste0("g", formatC(g, width = 3, flag = "0"))
sim_df$g <- as.factor(g)
## Prior predictions (methods c and d):
y_prior_pred <- f_generate_y(sim_df = ref_gr,
b_Intercept = subset(prior_r,
variable == "b_Intercept")$simulated_value,
b_x = subset(prior_r,
variable == "b_x")$simulated_value,
grouping_term = subset(prior_r,
variable == "sd_g__Intercept")$simulated_value,
sigma = subset(prior_r,
variable == "sigma")$simulated_value)
ref_gr$y_prior_pred <- y_prior_pred$y
rm(y_prior_pred)
y_prior_pred <- f_generate_y(sim_df = sim_df,
b_Intercept = subset(prior_r,
variable == "b_Intercept")$simulated_value,
b_x = subset(prior_r,
variable == "b_x")$simulated_value,
grouping_term = subset(prior_r,
variable == "sd_g__Intercept")$simulated_value,
sigma = subset(prior_r,
variable == "sigma")$simulated_value)
sim_df$y_prior_pred <- y_prior_pred$y
rm(y_prior_pred)
reference_c <- mean(ref_gr$y_prior_pred)
reference_d <- mean(sim_df$y_prior_pred)
b <- datasets$variables[r, ]
b <- as.numeric(b)
## Prior 'uncertainty' for ref_gr:
gamma <- sample(b[grep(x = colnames(datasets$variables),
pattern = "r_g[Group", fixed = T)],
size = 200, replace = T)
gamma <- matrix(gamma, nrow = 200, ncol = 1,
dimnames = list(levels(ref_gr$g), "Intercept"))
ref_gr$y_prior_pred_uncrtnty <- f_generate_y(sim_df = ref_gr,
b_Intercept =
b[which(colnames(datasets$variables) == "b_Intercept")],
b_x =
b[which(colnames(datasets$variables) == "b_x")],
grouping_term = gamma,
sigma =
b[which(colnames(datasets$variables) == "sigma")])$y
## Prior 'uncertainty' for sim_df:
gamma <- sample(b[grep(x = colnames(datasets$variables),
pattern = "r_g[Group", fixed = T)],
size = 20, replace = T)
gamma <- matrix(gamma, nrow = 20, ncol = 1,
dimnames = list(levels(sim_df$g), "Intercept"))
sim_df$y_prior_pred_uncrtnty <- f_generate_y(sim_df = sim_df,
b_Intercept =
b[which(colnames(datasets$variables) == "b_Intercept")],
b_x =
b[which(colnames(datasets$variables) == "b_x")],
grouping_term = gamma,
sigma =
b[which(colnames(datasets$variables) == "sigma")])$y
reference_e <- mean(ref_gr$y_prior_pred_uncrtnty)
reference_f <- mean(sim_df$y_prior_pred_uncrtnty)
B <- as.matrix(results$fits[[r]]) ## Posterior sample
posterior_equation_a <- exp(B[, "b_Intercept"] + 1 * B[, "b_x"])
posterior_equation_b <- exp(B[, "b_Intercept"] + 1 * B[, "b_x"] +
.5 * B[, "sd_g__Intercept"]^2)
Y_rep_ref_gr_g <- f_predict_brms_gaussian(results$fits[[r]], ref_gr)
Y_rep_ref_gr_u <- f_predict_brms_uncrtnty(results$fits[[r]], ref_gr)
Y_rep_sim_df_g <- f_predict_brms_gaussian(results$fits[[r]], sim_df)
Y_rep_sim_df_u <- f_predict_brms_uncrtnty(results$fits[[r]], sim_df)
stat_mean_ref_gr_uncrtnty_a <- stat_mean_ref_gr_gaussian_a <-
stat_mean_ref_gr_uncrtnty_b <- stat_mean_ref_gr_gaussian_b <-
stat_mean_sim_df_uncrtnty_a <- stat_mean_sim_df_gaussian_a <-
stat_mean_sim_df_uncrtnty_b <- stat_mean_sim_df_gaussian_b <-
##
stat_mean_ref_gr_uncrtnty_c <- stat_mean_ref_gr_gaussian_c <-
stat_mean_ref_gr_uncrtnty_d <- stat_mean_ref_gr_gaussian_d <-
stat_mean_sim_df_uncrtnty_c <- stat_mean_sim_df_gaussian_c <-
stat_mean_sim_df_uncrtnty_d <- stat_mean_sim_df_gaussian_d <-
##
stat_mean_ref_gr_uncrtnty_e <- stat_mean_ref_gr_gaussian_e <-
stat_mean_ref_gr_uncrtnty_f <- stat_mean_ref_gr_gaussian_f <-
stat_mean_sim_df_uncrtnty_e <- stat_mean_sim_df_gaussian_e <-
stat_mean_sim_df_uncrtnty_f <- stat_mean_sim_df_gaussian_f <-
##
eq_a_a <- eq_a_b <- eq_a_c <- eq_a_d <- eq_a_e <- eq_a_f <-
eq_b_a <- eq_b_b <- eq_b_c <- eq_b_d <- eq_b_e <- eq_b_f <-
rep(NA, nrow(Y_rep_ref_gr_g))
for (s in 1:nrow(Y_rep_ref_gr_g)) {
ref_gr$y_rep_gaussian <- Y_rep_ref_gr_g[s, ]
ref_gr$y_rep_uncrtnty <- Y_rep_ref_gr_u[s, ]
sim_df$y_rep_gaussian <- Y_rep_sim_df_g[s, ]
sim_df$y_rep_uncrtnty <- Y_rep_sim_df_u[s, ]
## Calculate stats:
stat_mean_ref_gr_gaussian_a[s] <- equation_a < mean(ref_gr$y_rep_gaussian)
stat_mean_ref_gr_uncrtnty_a[s] <- equation_a < mean(ref_gr$y_rep_uncrtnty)
stat_mean_ref_gr_gaussian_b[s] <- equation_b < mean(ref_gr$y_rep_gaussian)
stat_mean_ref_gr_uncrtnty_b[s] <- equation_b < mean(ref_gr$y_rep_uncrtnty)
stat_mean_sim_df_gaussian_a[s] <- equation_a < mean(sim_df$y_rep_gaussian)
stat_mean_sim_df_uncrtnty_a[s] <- equation_a < mean(sim_df$y_rep_uncrtnty)
stat_mean_sim_df_gaussian_b[s] <- equation_b < mean(sim_df$y_rep_gaussian)
stat_mean_sim_df_uncrtnty_b[s] <- equation_b < mean(sim_df$y_rep_uncrtnty)
##
stat_mean_ref_gr_gaussian_c[s] <- reference_c < mean(ref_gr$y_rep_gaussian)
stat_mean_ref_gr_uncrtnty_c[s] <- reference_c < mean(ref_gr$y_rep_uncrtnty)
stat_mean_ref_gr_gaussian_d[s] <- reference_d < mean(ref_gr$y_rep_gaussian)
stat_mean_ref_gr_uncrtnty_d[s] <- reference_d < mean(ref_gr$y_rep_uncrtnty)
stat_mean_sim_df_gaussian_c[s] <- reference_c < mean(sim_df$y_rep_gaussian)
stat_mean_sim_df_uncrtnty_c[s] <- reference_c < mean(sim_df$y_rep_uncrtnty)
stat_mean_sim_df_gaussian_d[s] <- reference_d < mean(sim_df$y_rep_gaussian)
stat_mean_sim_df_uncrtnty_d[s] <- reference_d < mean(sim_df$y_rep_uncrtnty)
##
stat_mean_ref_gr_gaussian_e[s] <- reference_e < mean(ref_gr$y_rep_gaussian)
stat_mean_ref_gr_uncrtnty_e[s] <- reference_e < mean(ref_gr$y_rep_uncrtnty)
stat_mean_ref_gr_gaussian_f[s] <- reference_f < mean(ref_gr$y_rep_gaussian)
stat_mean_ref_gr_uncrtnty_f[s] <- reference_f < mean(ref_gr$y_rep_uncrtnty)
stat_mean_sim_df_gaussian_e[s] <- reference_e < mean(sim_df$y_rep_gaussian)
stat_mean_sim_df_uncrtnty_e[s] <- reference_e < mean(sim_df$y_rep_uncrtnty)
stat_mean_sim_df_gaussian_f[s] <- reference_f < mean(sim_df$y_rep_gaussian)
stat_mean_sim_df_uncrtnty_f[s] <- reference_f < mean(sim_df$y_rep_uncrtnty)
##
eq_a_a[s] <- equation_a < posterior_equation_a[s]
eq_a_b[s] <- equation_b < posterior_equation_a[s]
eq_a_c[s] <- reference_c < posterior_equation_a[s]
eq_a_d[s] <- reference_d < posterior_equation_a[s]
eq_a_e[s] <- reference_e < posterior_equation_a[s]
eq_a_f[s] <- reference_f < posterior_equation_a[s]
##
eq_b_a[s] <- equation_a < posterior_equation_b[s]
eq_b_b[s] <- equation_b < posterior_equation_b[s]
eq_b_c[s] <- reference_c < posterior_equation_b[s]
eq_b_d[s] <- reference_d < posterior_equation_b[s]
eq_b_e[s] <- reference_e < posterior_equation_b[s]
eq_b_f[s] <- reference_f < posterior_equation_b[s]
}
## Calculate posterior predictive p-values:
p_stat_mean_ref_gr_gaussian_a[r] <- mean(stat_mean_ref_gr_gaussian_a)
p_stat_mean_ref_gr_gaussian_b[r] <- mean(stat_mean_ref_gr_gaussian_b)
p_stat_mean_ref_gr_gaussian_c[r] <- mean(stat_mean_ref_gr_gaussian_c)
p_stat_mean_ref_gr_gaussian_d[r] <- mean(stat_mean_ref_gr_gaussian_d)
p_stat_mean_ref_gr_gaussian_e[r] <- mean(stat_mean_ref_gr_gaussian_e)
p_stat_mean_ref_gr_gaussian_f[r] <- mean(stat_mean_ref_gr_gaussian_f)
##
p_stat_mean_ref_gr_uncrtnty_a[r] <- mean(stat_mean_ref_gr_uncrtnty_a)
p_stat_mean_ref_gr_uncrtnty_b[r] <- mean(stat_mean_ref_gr_uncrtnty_b)
p_stat_mean_ref_gr_uncrtnty_c[r] <- mean(stat_mean_ref_gr_uncrtnty_c)
p_stat_mean_ref_gr_uncrtnty_d[r] <- mean(stat_mean_ref_gr_uncrtnty_d)
p_stat_mean_ref_gr_uncrtnty_e[r] <- mean(stat_mean_ref_gr_uncrtnty_e)
p_stat_mean_ref_gr_uncrtnty_f[r] <- mean(stat_mean_ref_gr_uncrtnty_f)
##
p_stat_mean_sim_df_gaussian_a[r] <- mean(stat_mean_sim_df_gaussian_a)
p_stat_mean_sim_df_gaussian_b[r] <- mean(stat_mean_sim_df_gaussian_b)
p_stat_mean_sim_df_gaussian_c[r] <- mean(stat_mean_sim_df_gaussian_c)
p_stat_mean_sim_df_gaussian_d[r] <- mean(stat_mean_sim_df_gaussian_d)
p_stat_mean_sim_df_gaussian_e[r] <- mean(stat_mean_sim_df_gaussian_e)
p_stat_mean_sim_df_gaussian_f[r] <- mean(stat_mean_sim_df_gaussian_f)
##
p_stat_mean_sim_df_uncrtnty_a[r] <- mean(stat_mean_sim_df_uncrtnty_a)
p_stat_mean_sim_df_uncrtnty_b[r] <- mean(stat_mean_sim_df_uncrtnty_b)
p_stat_mean_sim_df_uncrtnty_c[r] <- mean(stat_mean_sim_df_uncrtnty_c)
p_stat_mean_sim_df_uncrtnty_d[r] <- mean(stat_mean_sim_df_uncrtnty_d)
p_stat_mean_sim_df_uncrtnty_e[r] <- mean(stat_mean_sim_df_uncrtnty_e)
p_stat_mean_sim_df_uncrtnty_f[r] <- mean(stat_mean_sim_df_uncrtnty_f)
##
p_stat_mean_eq_a_a[r] <- mean(eq_a_a)
p_stat_mean_eq_a_b[r] <- mean(eq_a_b)
p_stat_mean_eq_a_c[r] <- mean(eq_a_c)
p_stat_mean_eq_a_d[r] <- mean(eq_a_d)
p_stat_mean_eq_a_e[r] <- mean(eq_a_e)
p_stat_mean_eq_a_f[r] <- mean(eq_a_f)
##
p_stat_mean_eq_b_a[r] <- mean(eq_b_a)
p_stat_mean_eq_b_b[r] <- mean(eq_b_b)
p_stat_mean_eq_b_c[r] <- mean(eq_b_c)
p_stat_mean_eq_b_d[r] <- mean(eq_b_d)
p_stat_mean_eq_b_e[r] <- mean(eq_b_e)
p_stat_mean_eq_b_f[r] <- mean(eq_b_f)
if (r %% 50 < .5) {cat(".\n")} else {cat(".")} ## Progress 'bar'
}Organize results
results_holdout_ppc <- rbind(data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' = "exp(b0+b1)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_gaussian_a),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' = "exp(b0+b1)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_uncrtnty_a),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' = "exp(b0+b1+.5*sigma_gamma^2)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_gaussian_b),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' = "exp(b0+b1+.5*sigma_gamma^2)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_uncrtnty_b),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' = "exp(b0+b1)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_gaussian_a),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' = "exp(b0+b1)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_uncrtnty_a),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' = "exp(b0+b1+.5*sigma_gamma^2)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_gaussian_b),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' = "exp(b0+b1+.5*sigma_gamma^2)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_uncrtnty_b),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on ref. grid",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_gaussian_c),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on ref. grid",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_uncrtnty_c),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on sim. data",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_gaussian_d),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on sim. data",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_uncrtnty_d),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on ref. grid",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_gaussian_c),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on ref. grid",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_uncrtnty_c),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on sim. data",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_gaussian_d),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on sim. data",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_uncrtnty_d),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on ref. grid (uncertainty)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_gaussian_e),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on ref. grid (uncertainty)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_uncrtnty_e),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on sim. data (uncertainty)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_gaussian_f),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on sim. data (uncertainty)",
'posterior_predict_population' = "ref. grid",
'value' = p_stat_mean_ref_gr_uncrtnty_f),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on ref. grid (uncertainty)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_gaussian_e),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on ref. grid (uncertainty)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_uncrtnty_e),
data.frame('brms_sample_new_levels' = "gaussian",
'use_prior' =
"mean of prior pred. on sim. data (uncertainty)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_gaussian_f),
data.frame('brms_sample_new_levels' = "uncertainty",
'use_prior' =
"mean of prior pred. on sim. data (uncertainty)",
'posterior_predict_population' = "sim. data",
'value' = p_stat_mean_sim_df_uncrtnty_f),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1)]",
'use_prior' = "exp(b0+b1)",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_a_a),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1)]",
'use_prior' = "exp(b0+b1+.5*sigma_gamma^2)",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_a_b),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1+.5*sigma_gamma^2)]",
'use_prior' = "exp(b0+b1)",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_b_a),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1+.5*sigma_gamma^2)]",
'use_prior' = "exp(b0+b1+.5*sigma_gamma^2)",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_b_b),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1)]",
'use_prior' = "prior_pred_ref_gr_gaussian",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_a_c),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1+.5*sigma_gamma^2)]",
'use_prior' = "prior_pred_ref_gr_gaussian",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_b_c),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1)]",
'use_prior' = "prior_pred_sim_df_gaussian",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_a_d),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1+.5*sigma_gamma^2)]",
'use_prior' = "prior_pred_sim_df_gaussian",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_b_d),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1)]",
'use_prior' = "prior_pred_ref_gr_uncrtnty",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_a_e),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1+.5*sigma_gamma^2)]",
'use_prior' = "prior_pred_ref_gr_uncrtnty",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_b_e),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1)]",
'use_prior' = "prior_pred_sim_df_uncrtnty",
'posterior_predict_population' = "none",
'value' = p_stat_mean_eq_a_f),
data.frame('brms_sample_new_levels' =
"none [exp(b0+b1+.5*sigma_gamma^2)]",
'use_prior' = "prior_pred_sim_df_uncrtnty",
'posterior_predict_population' = "none",
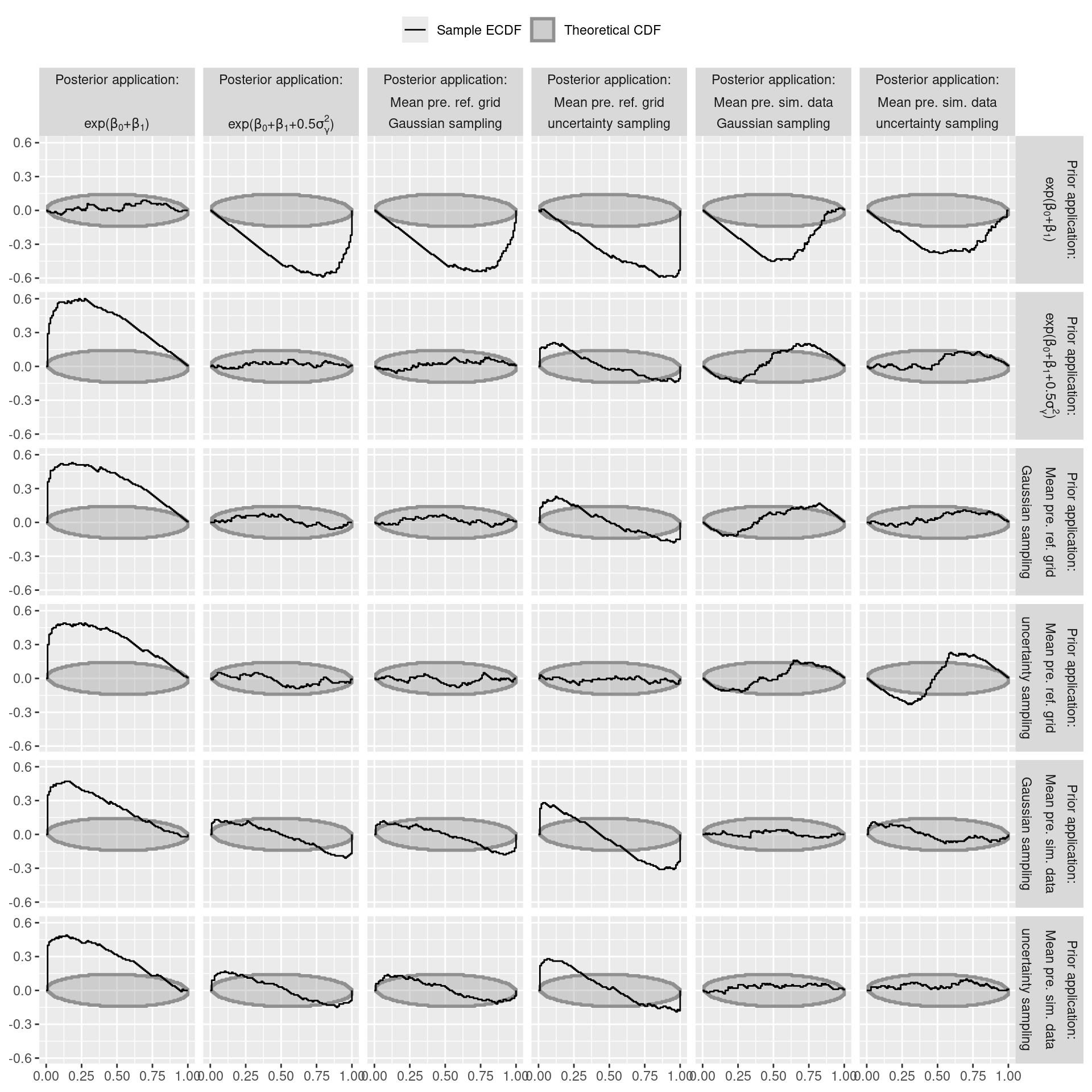
'value' = p_stat_mean_eq_b_f))Visualise QOI-Check
tmp <- results_holdout_ppc
tmp$variable <- paste0("Mean of prediction on ",
tmp$posterior_predict_population, " with\n",
tmp$brms_sample_new_levels, " sampling for gamma.\n",
"Prior application: ", tmp$use_prior)
tmp$variable <- gsub(x = tmp$variable, pattern = "gaussian",
replacement = "Gaussian", fixed = T)
tmp$rank <- tmp$value * 1000
tmp$max_rank <- 1000
tmp$sim_id <- rep(1:100, 6 * 6)
tmp <- dat_ecdf_diff(x = tmp)
tmp$ecdf_df$x1 <- sapply(X = strsplit(x = tmp$ecdf_df$variable, split = '\n',
fixed = T), FUN = function(x){x[1]})
tmp$ecdf_df$x2 <- sapply(X = strsplit(x = tmp$ecdf_df$variable, split = '\n',
fixed = T), FUN = function(x){x[2]})
tmp$ecdf_df$x3 <- sapply(X = strsplit(x = tmp$ecdf_df$variable, split = '\n',
fixed = T), FUN = function(x){x[3]})
tmp$ecdf_df$x3 <- gsub(x = tmp$ecdf_df$x3, pattern = "Prior application: ",
replacement = "", fixed = T)
tmp$ecdf_df$x3f <- as.factor(tmp$ecdf_df$x3)
tmp$ecdf_df$x4f <- as.factor(tmp$ecdf_df$x3)
levels(tmp$ecdf_df$x3f) <- c(expression("exp(" * beta[0] * "+" * beta[1] * ")"),
expression("exp(" * beta[0] * "+" * beta[1] *
"+0.5" * sigma[gamma]^2 * ")"),
expression(paste("Mean pre. ref. grid")),
expression(paste("Mean pre. ref. grid")),
expression(paste("Mean pre. sim. data")),
expression(paste("Mean pre. sim. data")),
expression(paste("Mean pre. ref. grid")),
expression(paste("Mean pre. ref. grid")),
expression(paste("Mean pre. sim. data")),
expression(paste("Mean pre. sim. data")))
levels(tmp$ecdf_df$x4f) <- c("",
"",
"Gaussian sampling",
"uncertainty sampling",
"Gaussian sampling",
"uncertainty sampling",
"Gaussian sampling",
"uncertainty sampling",
"Gaussian sampling",
"uncertainty sampling")
tmp$ecdf_df$x1a <- "Posterior application:"
tmp$ecdf_df$x3a <- "Prior application:"
tmp$ecdf_df$x1[tmp$ecdf_df$x1 == "Mean of prediction on none with"] <- " "
tmp$ecdf_df$x1f <- as.factor(tmp$ecdf_df$x1)
levels(tmp$ecdf_df$x1f) <- c(" ",
"Mean pre. ref. grid",
"Mean pre. sim. data")
tmp$ecdf_df$x2[tmp$ecdf_df$x2 == "none [exp(b0+b1)] sampling for gamma."] <-
"exp(b0+b1)"
tmp$ecdf_df$x2[tmp$ecdf_df$x2 ==
"none [exp(b0+b1+.5*sigma_gamma^2)] sampling for gamma."] <-
"exp(b0+b1+.5*sigma_gamma^2)"
tmp$ecdf_df$x2f <- as.factor(tmp$ecdf_df$x2)
levels(tmp$ecdf_df$x2f) <- c(expression("exp(" * beta[0] * "+" * beta[1] * ")"),
expression("exp(" * beta[0] * "+" * beta[1] *
"+0.5" * sigma[gamma]^2 * ")"),
expression(paste("Gaussian sampling")),
expression(paste("uncertainty sampling")))
col_color <- col_fill <-
c(colorspace::sequential_hcl(n = 20, pal = "Grays")[10], "black")
names(col_color) <- names(col_fill) <-
c("theoretical CDF", "sample ECDF")
l <- c('theoretical CDF' = expression("Theoretical CDF"),
'sample ECDF' = expression("Sample ECDF"))
ggplot(tmp$ecdf_df, aes(color = type, fill = type)) +
geom_ribbon(data = tmp$limits_df_trans,
aes(x = x, ymax = ymax, ymin = ymin), alpha = .33,
linewidth = 1) +
geom_step(aes(x = z, y = z_diff, group = variable)) +
scale_color_manual(name = "", values = col_color, labels = l) +
scale_fill_manual(name = "", values = col_fill, labels = l) +
scale_alpha_identity() +
xlab(NULL) +
ylab(NULL) +
facet_grid(cols = vars(x1a, x1f, x2f),
rows = vars(x3a, x3f, x4f),
labeller = labeller(x2f = label_parsed,
x3f = label_parsed)) +
theme(strip.text = element_text(margin = margin(b = 0, t = 0))) +
theme(legend.position = "top")