library("mgcv")
library("SBC")
library("brms")
library("cmdstanr")
library("ggplot2")
library("colorspace")
library("cowplot")
library("future")Intro
Relevant parts of the code given here is inspired by the resources of the great SBC package (Modrák et al., 2023) published at https://hyunjimoon.github.io/SBC/index.html.
Also, coding of posterior p-values is inspired by the split predictive check (Li & Huggins, 2024) R code published at https://github.com/TARPS-group/split-predictive-checks.
Software
We use the statistical software environment R (R Core Team, 2024), and R add-on packages SBC (Modrák et al., 2023), brms (Bürkner, 2017, 2018, 2021), mgcv (Wood, 2011), cmdstanr (Gabry et al., 2024), ggplot2 (Wickham, 2016), colorspace (Stauffer et al., 2009), and future (Bengtsson, 2021).
This document is produced using Quarto (Allaire et al., 2024).
Organize R Session
Load packages:
Settings:
rm(list = ls())
options(brms.backend = "cmdstanr")
# Using parallel processing
plan(multisession)
options(SBC.min_chunk_size = 5)
# Setup caching of results
cache_dir <- "./SBC_cache"
if(!dir.exists(cache_dir)) {
dir.create(cache_dir)
}Data preparation function for graphical uniformity checks:
# convenient wrapper around mgcv::PredictMat
PredictMat <- function(object, data, ...) {
data <- rm_attr(data, "terms")
out <- mgcv::PredictMat(object, data = data, ...)
if (length(dim(out)) < 2L) {
# fixes issue #494
out <- matrix(out, nrow = 1)
}
out
}
# convenient wrapper around mgcv::smoothCon
smoothCon <- function(object, data, ...) {
data <- rm_attr(data, "terms")
vars <- setdiff(c(object$term, object$by), "NA")
for (v in vars) {
if (is_like_factor(data[[v]])) {
# allow factor-like variables #562
data[[v]] <- as.factor(data[[v]])
} else if (inherits(data[[v]], "difftime")) {
# mgcv cannot handle 'difftime' variables
data[[v]] <- as.numeric(data[[v]])
}
}
mgcv::smoothCon(object, data = data, ...)
}
# Aid prediction from smooths represented as 'type = 2'
# code obtained from the doc of ?mgcv::smooth2random
# @param sm output of mgcv::smoothCon
# @param re output of mgcv::smooth2random
# @param data new data supplied for prediction
# @return A list of the same structure as returned by mgcv::smooth2random
s2rPred <- function(sm, re, data) {
#sm = sm[[1]] ## for check!
#re = hmmmm ## for check!
#data = nd ## for check!
# prediction matrix for new data
X <- mgcv::PredictMat(sm, data)
# transform to RE parameterization
if (!is.null(re$trans.U)) {
X <- X %*% re$trans.U
}
if (is.null(re$trans.D)) {
# regression spline without penalization
out <- list(Xf = X)
} else {
X <- t(t(X) * re$trans.D)
# re-order columns according to random effect re-ordering
X[, re$rind] <- X[, re$pen.ind != 0]
# re-order penalization index in same way
pen.ind <- re$pen.ind
pen.ind[re$rind] <- pen.ind[pen.ind > 0]
# start returning the object
Xf <- X[, which(re$pen.ind == 0), drop = FALSE]
out <- list(rand = list(), Xf = Xf)
for (i in seq_along(re$rand)) {
# loop over random effect matrices
out$rand[[i]] <- X[, which(pen.ind == i), drop = FALSE]
attr(out$rand[[i]], "s.label") <- attr(re$rand[[i]], "s.label")
}
names(out$rand) <- names(re$rand)
}
out
}Data preparation function for graphical uniformity checks:
## from https://github.com/TeemuSailynoja/simultaneous-confidence-bands/tree/main
dat_ecdf_diff <- function (x, variables = NULL, K = NULL,
gamma = NULL, prob = 0.95,
combine_variables = NULL,
ecdf_alpha = NULL, ...,
parameters = NULL) {
if (!is.null(parameters)) {
warning("The `parameters` argument is deprecated use `variables` instead.")
if (is.null(variables)) {
variables <- parameters
}
}
ecdf_data <- data_for_ecdf_plots(x, variables = variables,
prob = prob, K = K, gamma = gamma,
combine_variables = combine_variables,
ecdf_alpha = ecdf_alpha, ...)
if (ecdf_data$N < 50 && is.null(K)) {
message("With less than 50 simulations, we recommend using plot_ecdf as it has better fidelity.\n",
"Disable this message by explicitly setting the K parameter. ",
"You can use the strings \"max\" (high fidelity) or \"min\" (nicer plot) or choose a specific integer.")
}
N <- ecdf_data$N
K <- ecdf_data$K
z <- ecdf_data$z
ecdf_df <- dplyr::mutate(ecdf_data$ecdf_df, z_diff = ecdf -
z, type = "sample ECDF")
limits_df_trans <- dplyr::mutate(ecdf_data$limits_df,
ymax = upper - uniform_val,
ymin = lower - uniform_val,
type = "theoretical CDF")
return(list('ecdf_df' = ecdf_df,
'limits_df_trans' = limits_df_trans))
}Data Generator
f_generate_y <- function(sim_df,
eta,
sigma) {
sim_df$'mu_y' <- eta
sim_df$'sigma_y' <- rep(sigma, nrow(sim_df))
sim_df$'y' <- rnorm(n = nrow(sim_df),
mean = sim_df$'mu_y',
sd = sim_df$'sigma_y')
return(sim_df)
}
f_generate_xz <- function(N, b_Intercept, phi) {
mu <- plogis(b_Intercept)
return(rbeta(n = N, shape1 = mu * phi, shape2 = (1 - mu) * phi))
}
f_generate_one_sim <- function(N) {
prior_mu <- list('b_x_Intercept' = 0,
'b_z_Intercept' = -1.1,
'b_y_Intercept' = .5,
'bs_y_sxz_1' = 1,
'bs_y_sxz_2' = -1,
'sds_y_sxz_1' = 1,
'phi_x' = 3,
'phi_z' = 3,
'sigma_y' = .1)
prior_sd <- list('b_x_Intercept' = .001,
'b_z_Intercept' = .001,
'b_y_Intercept' = .1,
'bs_y_sxz_1' = .1,
'bs_y_sxz_2' = .1,
'sds_y_sxz_1' = .1,
'phi_x' = .001,
'phi_z' = .001,
'sigma_y' = .01)
b_y_Intercept <- rnorm(n = 1,
mean = prior_mu$b_y_Intercept,
sd = prior_sd$b_y_Intercept)
bs_y_sxz_1 <- rnorm(n = 1,
mean = prior_mu$bs_y_sxz_1,
sd = prior_sd$bs_y_sxz_1)
bs_y_sxz_2 <- rnorm(n = 1,
mean = prior_mu$bs_y_sxz_2,
sd = prior_sd$bs_y_sxz_2)
sds_y_sxz_1 <- rnorm(n = 1,
mean = prior_mu$sds_y_sxz_1,
sd = prior_sd$sds_y_sxz_1)
sds_y_sxz_1 <- abs(sds_y_sxz_1)
sigma_y <- rnorm(n = 1,
mean = prior_mu$sigma_y,
sd = prior_sd$sigma_y)
b_x_Intercept <- rnorm(n = 1,
mean = prior_mu$b_x_Intercept,
sd = prior_sd$b_x_Intercept)
phi_x <- rnorm(n = 1,
mean = prior_mu$phi_x,
sd = prior_sd$phi_x)
phi_x <- abs(phi_x)
b_z_Intercept <- rnorm(n = 1,
mean = prior_mu$b_z_Intercept,
sd = prior_sd$b_z_Intercept)
phi_z <- rnorm(n = 1,
mean = prior_mu$phi_z,
sd = prior_sd$phi_z)
phi_z <- abs(phi_z)
x <- f_generate_xz(N, b_x_Intercept, phi_x)
z <- f_generate_xz(N, b_z_Intercept, phi_z)
dat_xz <- data.frame('x' = x, 'z' = z)
sm <- mgcv::smoothCon(object = s(x, z, k = 10), data = dat_xz,
absorb.cons = TRUE, modCon = 3, diagonal.penalty = TRUE)
sm2r <- mgcv::smooth2random(object = sm[[1]], vnames = c('x', 'z'), type = 2)
Xf <- sm2r[['Xf']]
Xr <- sm2r[['rand']][['Xr']]
s_y_sxz <- t(MASS::mvrnorm(n = 1, mu = rep(0, ncol(Xr)),
Sigma = sds_y_sxz_1 * diag(ncol(Xr))))
variables <- list('b_y_Intercept' = b_y_Intercept,
'bs_y_sxz_1' = bs_y_sxz_1,
'bs_y_sxz_2' = bs_y_sxz_2,
'sds_y_sxz_1' = sds_y_sxz_1,
's_y_sxz[1]' = s_y_sxz[1, 1],
's_y_sxz[2]' = s_y_sxz[1, 2],
's_y_sxz[3]' = s_y_sxz[1, 3],
's_y_sxz[4]' = s_y_sxz[1, 4],
's_y_sxz[5]' = s_y_sxz[1, 5],
's_y_sxz[6]' = s_y_sxz[1, 6],
's_y_sxz[7]' = s_y_sxz[1, 7],
'sigma_y' = sigma_y,
'b_x_Intercept' = b_x_Intercept,
'b_z_Intercept' = b_z_Intercept,
'phi_x' = phi_x,
'phi_z' = phi_z)
etaf <- Xf %*% c(bs_y_sxz_1, bs_y_sxz_2)
etar <- Xr %*% t(s_y_sxz)
eta <- b_y_Intercept + etaf + etar
return(list('variables' = variables,
'generated' = f_generate_y(sim_df = dat_xz,
eta = eta,
sigma = sigma_y)))
}
f_generate_n_sims <- SBC_generator_function(f_generate_one_sim,
N = 1000)
set.seed(123)
datasets <- generate_datasets(f_generate_n_sims, 20)
head(datasets$generated[[1]]) x z mu_y sigma_y y
1 0.6428125 0.471219239 0.3496019 0.1012929 0.33045945
2 0.1542820 0.158118671 0.1796588 0.1012929 0.10296001
3 0.3846568 0.116883626 0.1478711 0.1012929 0.22810111
4 0.3718601 0.228222288 0.2942474 0.1012929 0.43050453
5 0.4644184 0.003815211 0.1276784 0.1012929 0.05732729
6 0.6761364 0.082005730 1.0524761 0.1012929 1.00740764Backend
priors <- prior(normal(.5,.1), class = "Intercept", resp = "y") +
prior(normal(1,.1), class = "b", coef = "sxz_1", resp = "y") +
prior(normal(-1,.1), class = "b", coef = "sxz_2", resp = "y") +
prior(normal(1,.1), class = "sds", resp = "y") +
prior(normal(.1,.01), class = "sigma", resp = "y") +
prior(normal(0,.001), class = "Intercept", resp = "x") +
prior(normal(-1.1,.001), class = "Intercept", resp = "z") +
prior(normal(3,.001), class = "phi", resp = "x") +
prior(normal(3,.001), class = "phi", resp = "z")
frmla <- bf(x ~ 1, family = Beta(link = "logit")) +
bf(z ~ 1, family = Beta(link = "logit")) +
bf(y ~ s(x, z, k = 10), family = gaussian())
f_backend <- SBC_backend_brms(frmla,
prior = priors,
chains = 1,
iter = 2000,
template_data = datasets$generated[[1]],
out_stan_file = file.path(cache_dir,
"case_study_II.stan"))set.seed(0)
results <- compute_SBC(datasets = datasets,
backend = f_backend,
cache_mode = "results",
cache_location = file.path(cache_dir,
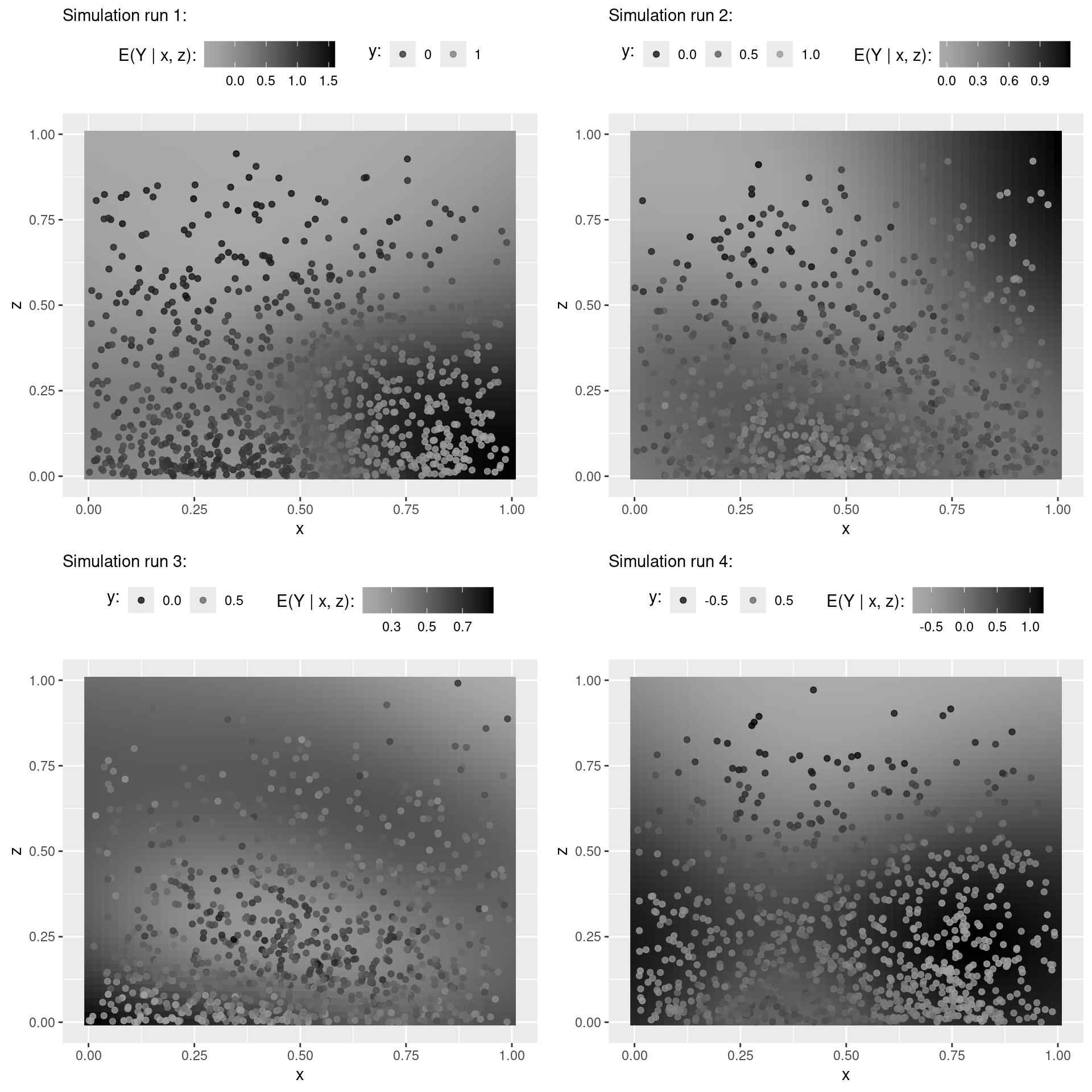
"SBC_results_case_study_II"))Visualize bivariate smooth at first four simulation runs
nd <- expand.grid('x' = seq(0, 1, by = .02), 'z' = seq(0, 1, by = .02))
p_list <- vector("list", 4)
for (r in 1:4) {
m <- results$fits[[r]]
fit <- fitted(object = m, newdata = nd)
nd$fit_y <- fit[, 'Estimate', 'y']
p_list[[r]] <- ggplot(data = nd, aes(x = x, y = z)) +
geom_raster(aes(fill = fit_y)) +
scale_fill_continuous_sequential(pal = "Grays", l1 = 0, l2 = 70) +
geom_point(data = m$data, aes(color = y), alpha = .8) +
scale_color_continuous_sequential(breaks = scales::breaks_extended(n = 3),
pal = "Grays", l1 = 0, l2 = 70, rev = FALSE) +
# colorspace::scale_color_continuous_sequential(pal = "Rocket") +
guides(color = guide_legend(nrow = 1)) +
labs(color = "y:", fill = "E(Y | x, z):",
subtitle = paste0("Simulation run ", r, ":")) +
theme(legend.position = 'top') +
theme(legend.title = element_text(vjust = .8))
}
cowplot::plot_grid(plotlist = p_list, ncol = 2)QOI-Check
f_postprocessing <- function(m, b, focus_on = 'x', at = .8,
integration_grid = seq(.025, .975, by = .05)) {
if (!(focus_on %in% c('x', 'z'))) {
stop("'focus_on' must be either 'x' or 'z'.")
}
nd <- data.frame('variable1' = at)
nd <- as.data.frame(expand.grid('variable1' = nd$variable1,
'variable2' = integration_grid))
names(nd) <- c('x', 'z')
fit <- fitted(m, newdata = nd, dpar = "mu", summary = F)
if (focus_on == 'x') {
phi <- fitted(m, newdata = nd, resp = "z", dpar = "phi", summary = F)
shape1 <- fit[, , "z"] * phi
shape2 <- (1 - fit[, , "z"]) * phi
d <- shape1 * 0
for (i in 1:nrow(nd)) {
d[, i] <- dbeta(x = nd$z[i], shape1 = shape1[, i], shape2 = shape2[, i])
}
}
if (focus_on == 'z') {
phi <- fitted(m, newdata = nd, resp = "x", dpar = "phi", summary = F)
shape1 <- fit[, , "x"] * phi
shape2 <- (1 - fit[, , "x"]) * phi
d <- shape1 * 0
for (i in 1:nrow(nd)) {
d[, i] <- dbeta(x = nd$x[i], shape1 = shape1[, i], shape2 = shape2[, i])
}
}
d <- d / apply(d, MAR = 1, FUN = sum)
tmp <- abs(apply(d, MAR = 1, FUN = sum) - 1)
if (any(tmp > 1e-10)) {
stop("No proper calculation of weights!")
}
return(list('Weighted' = apply(d * fit[, , 'y'], MAR = 1, FUN = sum),
'Unweighted' = apply(fit[, , 'y'], MAR = 1, FUN = mean)))
}
ranks_rgpp <- NULL
for (j in 1:20) {
cat(paste0("j = ", j, ":\n"))
cat(" f_postprocessing_fast():\n")
cat(" ")
res <- all_weighted <- NULL
x_grid <- seq(.025, .975, by = .05)
for (i in 1:length(x_grid)) {
x <- x_grid[i]
tmp <- f_postprocessing(results$fits[[j]],
as.data.frame(datasets$variables)[j, ],
focus_on = 'x', x,
seq(.0005, .9995, by = .001))
all_weighted <- cbind(all_weighted, tmp[['Weighted']])
for (method in c('Weighted', 'Unweighted')) {
res <- rbind(res,
data.frame('x' = x,
'y' = mean(tmp[[method]]),
'method' = method,
'stat' = 'Mean'))
res <- rbind(res,
data.frame('x' = x,
'y' = median(tmp[[method]]),
'method' = method,
'stat' = 'Median'))
res <- rbind(res,
data.frame('x' = x,
'y' = quantile(tmp[[method]], probs = .025),
'method' = method,
'stat' = '2.5% quantile'))
res <- rbind(res,
data.frame('x' = x,
'y' = quantile(tmp[[method]], probs = .975),
'method' = method,
'stat' = '97.5% quantile'))
}
cat(".")
}
res0 <- res
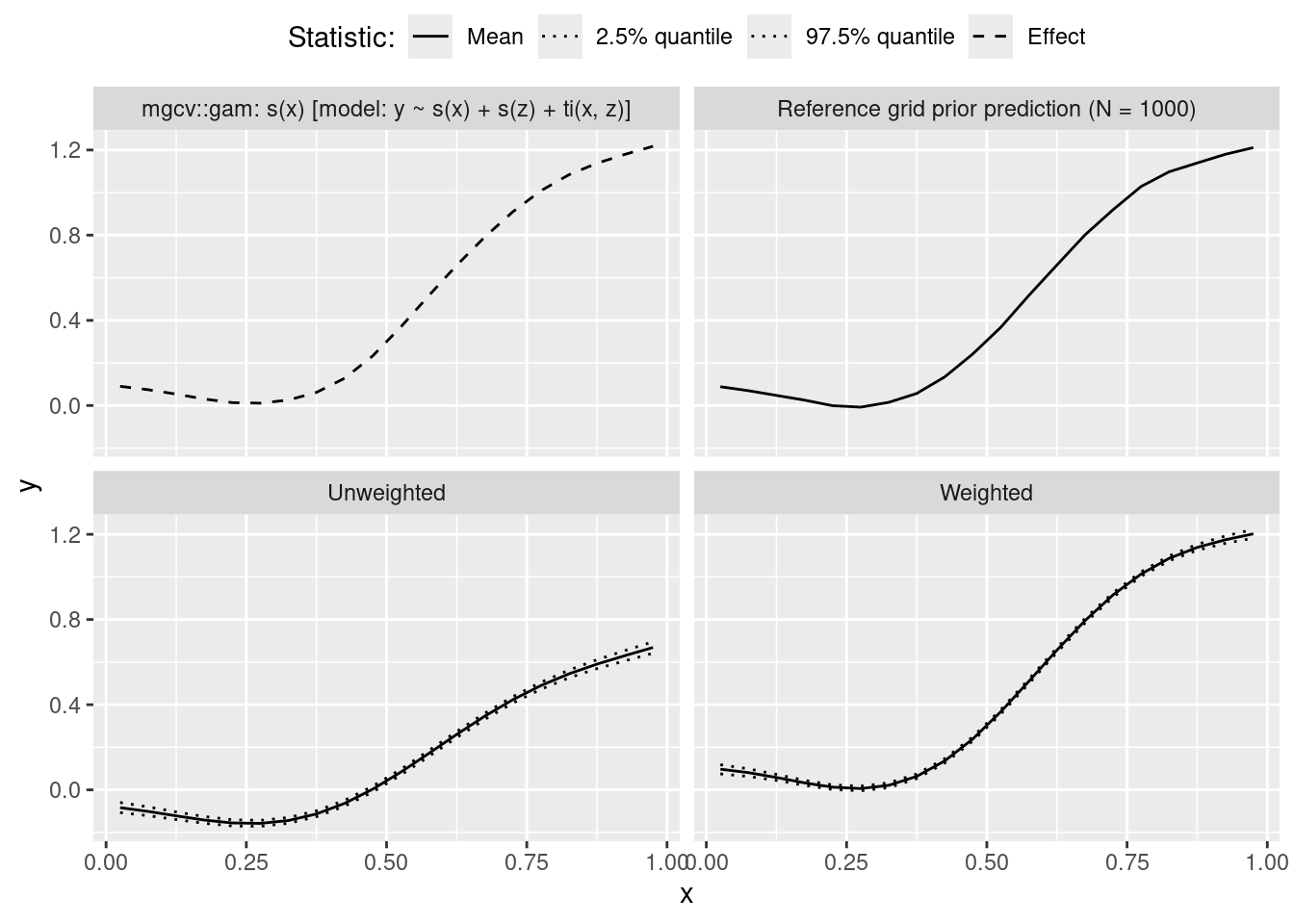
## Compare to GAM s() + s() + ti()
## https://cran.r-project.org/web/packages/mgcv/mgcv.pdf
## p. 79:
## "Sometimes it is interesting to specify smooth models with a main
## effects + interaction structure such as
## E(yi) = f1(xi) + f2(zi) + f3(xi, zi)
## [...]
## for example.
## Such models should be set up using ti terms in the model formula.
## For example:
## y ~ ti(x) + ti(z) + ti(x,z),
## [...]
## The ti terms produce interactions with the component main effects
## excluded appropriately. (There is in fact no need to use ti terms
## for the main effects here, s terms could also be used.)" ## "[...] in fact no need to use ti terms for the main effects here,
## s terms could also be used."
m_gam <- gam(y ~ s(x) + s(z) + ti(x, z), data = datasets$generated[[j]])
nd <- data.frame(x = x_grid, z = x_grid)
tmp <- predict(m_gam, type = "terms", newdata = nd)
res <- rbind(res,
data.frame(x = x_grid,
y = tmp[, 's(x)'] + coef(m_gam)[1],
method = "mgcv::gam: s(x) [model: y ~ s(x) + s(z) + ti(x, z)]",
stat = "Effect"))
## ############################################
## Reference grid posterior predictive check ##
## ############################################
cat("\n")
cat(" reference grid prior prediction:\n")
cat(" ")
x_grid <- seq(.025, .975, by = .05)
ref_grid_prior_mean_y_x <- rep(NA, length(x_grid))
for (i in 1:length(x_grid)) {
b <- as.data.frame(datasets$variables)[j, ]
z <- f_generate_xz(N = 10000,
b_Intercept = as.numeric(b['b_z_Intercept']),
phi = as.numeric(b['phi_z']))
dat_xz <- data.frame('x' = x_grid[i], 'z' = z)
sm <- mgcv::smoothCon(object = s(x, z, k = 10), data = results$fits[[j]]$data,
absorb.cons = TRUE, modCon = 3, diagonal.penalty = TRUE)
sm2r <- mgcv::smooth2random(object = sm[[1]], vnames = c('x', 'z'), type = 2)
sm2r_pre <- s2rPred(sm = sm[[1]], re = sm2r, data = dat_xz)
Xf <- sm2r_pre[['Xf']]
Xr <- sm2r_pre[['rand']][['Xr']]
etaf <- Xf %*% as.numeric(b[grep(names(b), pattern = "bs_", fixed = T)])
etar <- Xr %*% as.numeric(b[grep(names(b), pattern = "s_y_sxz[", fixed = T)])
eta <- as.numeric(b['b_y_Intercept']) + etaf + etar
dat_xz <- f_generate_y(dat_xz, as.numeric(eta), as.numeric(b["sigma_y"]))
ref_grid_prior_mean_y_x[i] <- mean(dat_xz$y)
cat(".")
}
cat("\n")
res <- rbind(res,
data.frame(x = x_grid,
y = ref_grid_prior_mean_y_x,
method = "Reference grid prior prediction (N = 1000)",
stat = "Mean"))
res$stat <- factor(res$stat, levels = c("Mean", "Median", "2.5% quantile",
"97.5% quantile", "Effect"))
if (j == 1) {
p <- ggplot(data = subset(res, stat != "Median"), aes(x = x, y = y)) +
geom_line(aes(group = paste0(stat, method),
linetype = stat)) + #, color = method
theme(legend.position = "top", legend.box="vertical", legend.margin=margin()) +
guides(color = "none") +
scale_linetype_manual(values = c(1, 3, 3, 2)) +
facet_wrap(~ method) +
labs(color = "Method:", linetype = "Statistic:")
}
rgpp_all_weighted <- rbind(subset(res,
method == "reference grid prior prediction (N = 1000)")$y,
all_weighted)
ranks_rgpp <- cbind(ranks_rgpp,
apply(rgpp_all_weighted, MAR = 2,
FUN = function(x){mean(x[-1] > x[1])}))
}
ranks <- data.frame('rank' = reshape2::melt(ranks_rgpp)$value,
'max_rank' = 1,
'variable' = reshape2::melt(ranks_rgpp)$Var1,
'sim_id' = reshape2::melt(ranks_rgpp)$Var2)
ranks$rank <- ranks$rank * 1000
ranks$rank <- ranks$rank - 1 + runif(nrow(ranks))
ranks$max_rank <- ranks$max_rank * 1000
ranks$variable <- factor(paste("x:", x_grid[ranks$variable]),
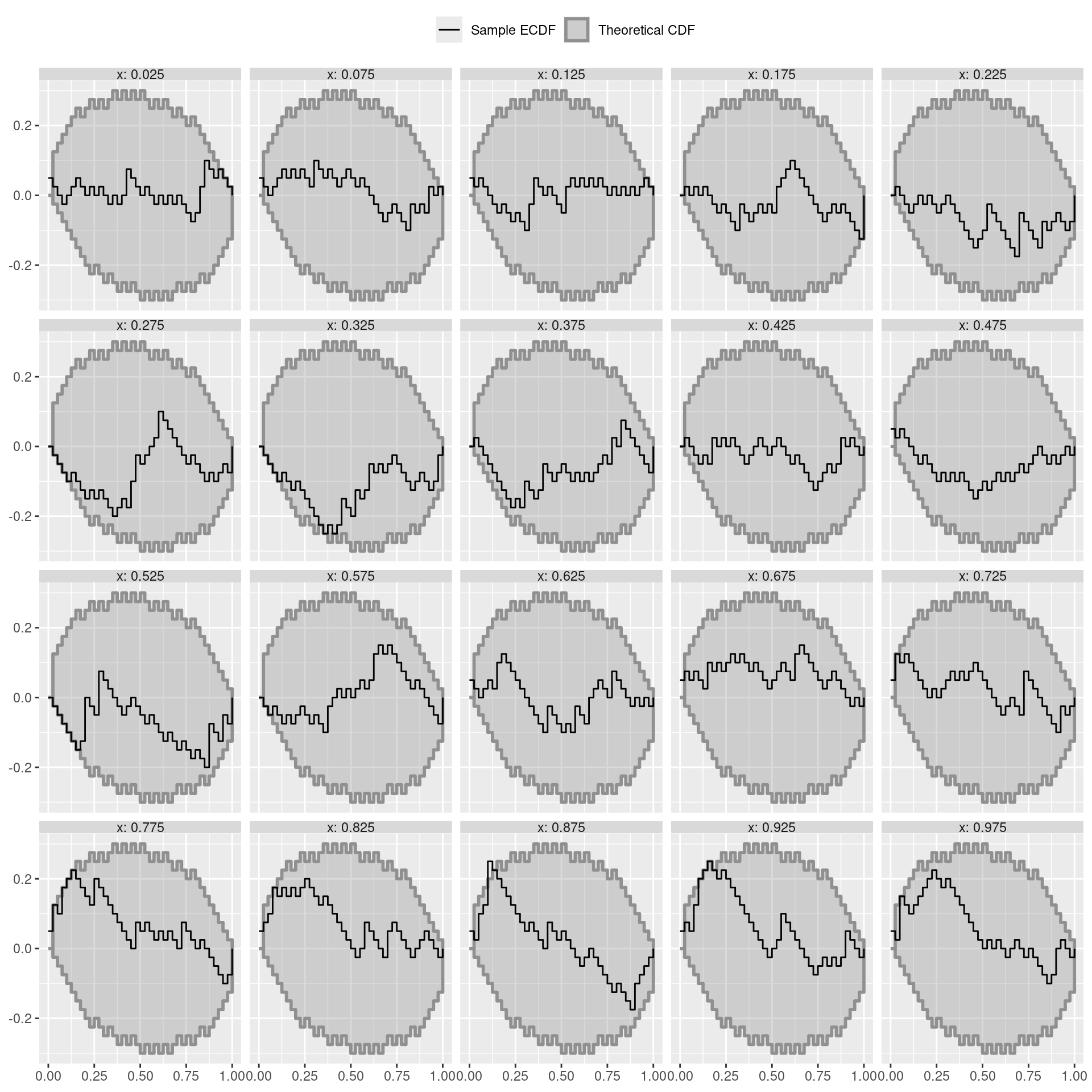
levels = paste("x:", x_grid))tmp <- dat_ecdf_diff(ranks, variables = levels(ranks$variable), K = 40)
col_color <- c(colorspace::sequential_hcl(n = 20, pal = "Grays")[10], "black")
col_fill <- c(colorspace::sequential_hcl(n = 20, pal = "Grays")[10], "black")
names(col_color) <- names(col_fill) <-
c("theoretical CDF", "sample ECDF")
l <- c('theoretical CDF' = expression("Theoretical CDF"),
'sample ECDF' = expression("Sample ECDF"))
ggplot(tmp$ecdf_df, aes(color = type, fill = type)) +
geom_ribbon(data = tmp$limits_df_trans,
aes(x = x, ymax = ymax, ymin = ymin), alpha = .33,
linewidth = 1) +
geom_step(aes(x = z, y = z_diff, group = variable)) +
scale_color_manual(name = "", values = col_color, labels = l) +
scale_fill_manual(name = "", values = col_fill, labels = l) +
scale_alpha_identity() +
xlab(NULL) +
ylab(NULL) +
facet_wrap(~variable) +
theme(strip.text = element_text(margin = margin(b = 0, t = 0))) +
theme(legend.position = "top")